
| Size | Price | Stock | Qty |
|---|---|---|---|
| 25mg |
|
||
| 50mg |
|
||
| 100mg |
|
||
| 250mg |
|
||
| 500mg |
|
||
| Other Sizes |
|
Purity: ≥98%
Tariquidar methanesulfonate hydrate (formerly also known as XR9576) is a novel potent and selective noncompetitive inhibitor of P-glycoprotein (P-gp) with Kd of 5.1 nM in CHrB30 cell line, it reverses drug resistance in MDR cell Lines. Tariquidar is currently undergoing research as an adjuvant against multidrug resistance in cancer. Tariquidar non-competitively binds to the p-glycoprotein transporter, thereby inhibiting transmembrane transport of anticancer drugs. Inhibition of transmembrane transport may result in increased intracellular concentrations of an anticancer drug, thereby augmenting its cytotoxicity.
| Targets |
P-gp (Kd = 5.1 nM)[1]
|
||
|---|---|---|---|
| ln Vitro |
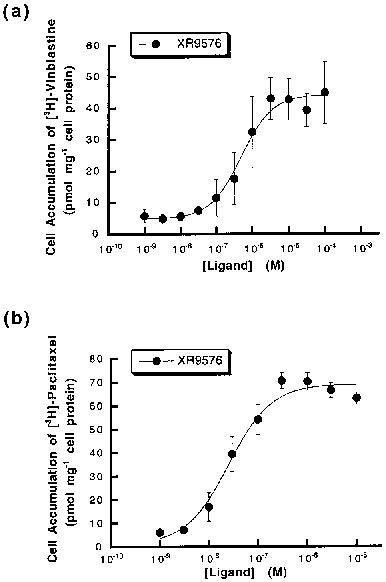
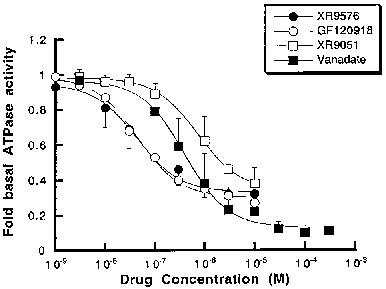
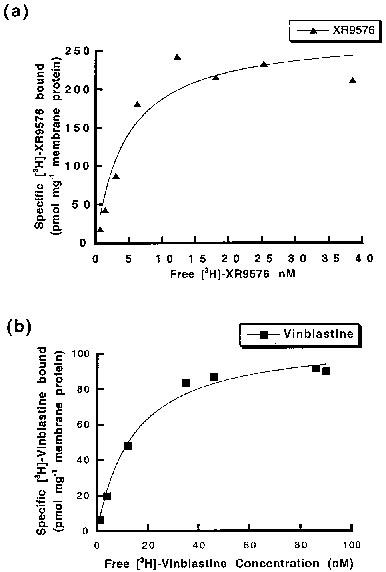
Tariquidar methanesulfonate, hydrate (XR9576 methanesulfonate, hydrate) raises the steady-state accumulation of these cytotoxics in CHrB30 cells to levels seen in non-P-gp-expressing AuxB1 cells (EC50=487±50 nM), making it a potent modulator of P-gp mediated [3H]-Vinblastine and [3H]-Paclitaxel transport. [3H]-Tariquidar has the strongest affinity (Kd=5.1±0.9 nM, n=7) and the maximum binding capacity (Bmax) of 275±15 pmol/mg membrane protein when it comes to CHrB30 membranes. Unlike the ancestral cell line, the modulators XR9576 (EC50=487±50 nM) increase the accumulation of [3H]-Vinblastine in a dose-dependent manner. With a strong IC50 value of 43±9 nM, the MDR modulator Tariquidar can inhibit 60–70% of the activity of the vanadate-sensitive ATPase[1]. Tariquidar (XR9576) enhances the cytotoxicity of several medications, including as Vincristine, Etoposide, Paclitaxel, and Doxorubicin; in the presence of 25–80 nM Tariquidar, resistance is completely reversed. Strong photoaffinity labeling of P-gp by [3H]Azidopine is inhibited by tariquidar, suggesting a direct interaction with the protein[2].
|
||
| ln Vivo |
Additionally, coadministration of Tariquidar (6–12 mg/kg po) fully restores the antitumor activity of Paclitaxel, Etoposide, and Vincristine against two highly resistant MDR human tumor xenografts (2780AD, H69/LX4) in nude mice. These mice bear the intrinsically resistant MC26 colon tumors, and coadministration of Tariquidar methanesulfonate, hydrate (XR9576 methanesulfonate, hydrate) potentiates the antitumor activity of Doxorubicin without a significant increase in toxicity. It has also been discovered that tariquidar dramatically increases the antitumor activity of doxorubicin against sc MC26 tumors in vivo[2].
|
||
| Enzyme Assay |
ATP hydrolytic activity of P-gp in CHrB30 membranes[1]
A previously described colorimetric assay was used to measure inorganic phosphate liberation following ATP hydrolysis (Chifflet et al., 1988). Membranes (1 μg protein) were incubated with Na2ATP (2 mm) in a total assay volume of 50 μl in buffer containing (mm): Tris pH 7.4 50, MgSO4 5, 0.02% NaN3, NH4Cl 150 for 20 min at 37°C. The ATPase activity was linear to 40 min at 37°C. Modulators (from DMSO stocks) and the ATPase inhibitor vanadate, were added in the concentration range 10−9–10–5 m. The final DMSO concentration was always <1%, a level known not to alter ATPase activity. The effect of drugs on the ATPase activity was fitted by the general dose-response relationship (see above). Specific drug binding to P-glycoprotein[1] A rapid filtration assay was used to measure the binding of [3H]-vinblastine, [3H]-paclitaxel and [3H]-XR9576 to P-gp in CHrB30 membranes as previously described (Ferry et al., 1992). Membranes were incubated with appropriate radioligand in a total buffer volume of 200 μl (50 mm Tris pH 7.4) for a period of 2–3 h to reach equilibrium. Washing buffer (3 ml) containing 20 mm MgSO4, 20 mm Tris (pH 7.4) was then added and the samples filtered under vacuum through a single GF/F filter in a filtration manifold to separate bound and free ligand. After further washing (2×3 ml) the amount of bound ligand was determined by liquid scintillation counting. Non-specific binding was defined as the amount of [3H]-ligand bound in the presence of at least a 100 fold excess of competing ligand (indicated in Results) and was subtracted from all values. Determination of the capacity and affinity of [3H]-ligand binding was achieved by saturation isotherm analysis. Membranes were incubated with increasing concentration of labelled drug and the amount bound (pmol mg−1) plotted as a function of free ligand concentration. |
||
| Cell Assay |
Cell culture[1]
The Chinese hamster ovary parental (sensitive) AuxB1 and the resistant CHrB30 cells were grown as previously described in α-minimum essential medium (α-MEM) containing 10% foetal calf serum (Kartner et al., 1983). The CHrB30 cells, derived from AuxB1 cells by step wise selection in colchicine (Kartner et al., 1983), express P-gp and selection pressure was maintained by supplementing media with 30 μg ml−1 colchicine. Plasma membrane preparation[1] Plasma membranes were prepared following disruption of CHrB30 cells using nitrogen cavitation and collection with sucrose density centrifugation as previously described (Lever, 1977). The final membrane preparation was stored at −70°C, at protein concentrations of 5–10 mg ml–1 in buffer containing 0.25 m sucrose, 10 mm Tris HCI (pH 7.5) and including the protease inhibitors leupeptin (0.1 mg ml−1), pepstatin A (0.1 mg ml−1) and benzamidine (1 mm). Steady-state drug accumulation assay[1] AuxB1 and CHrB30 cells were grown to confluency in 12-well (24 mm) tissue culture dishes and the steady-state accumulation of [3H]-vinblastine was measured as previously described (Martin et al., 1997). Accumulation was initiated by the addition of 0.1 μCi [3H]-vinblastine and unlabelled vinblastine to a final concentration of 100 nm. The accumulation of [3H]-paclitaxel was measured using 0.1 μCi [3H]-paclitaxel and unlabelled drug to a final concentration of 1 μm. Cells were incubated in a reaction volume of 1 ml for 60 min at 37°C under 5% CO2 in order to reach steady-state. The effect of the modulators XR9576 and GF120918 on [3H]-ligand accumulation was investigated in the concentration range 10−9–10−6 m. Modulators were added from a DMSO stock giving a final solvent concentration of 0.2% (v v−1). Following cell harvesting, accumulated drug was measured by liquid scintillation counting and normalized for cell protein content. Plots of amount accumulated as a function of modulator concentration were fitted with the general dose-response equation (De Lean et al., 1978). The accumulation of [3H]-XR9576 was also measured in AuxB1 and CHrB30 cells using several concentrations of radiolabelled drug (1–300 nm) in the presence and absence of 1 μm GF120918 and followed over a 60 min period, as described above. |
||
| Animal Protocol |
|
||
| References |
|
||
| Additional Infomation |
1 The kinetics and nature of equilibrium binding were used to characterize the molecular interaction of the anthranilic acid derivative [3H]-XR9576 with the multidrug resistance P-glycoprotein (P-gp). XR9576 displayed specific high-affinity binding to P-gp (Bmax = 275 pmol mg-1, Kd = 5.1 nM). The transport substrates [3H]-vinblastine and [3H]-paclitaxel displayed 4 fold and 20 fold lower affinity respectively for P-gp. The duration of action of XR9576 with P-gp was increased in comparison to that of vinblastine which displayed a slower rate of association and a faster dissociation rate. 2 The relative affinities of several modulators and transport substrates to interact with P-gp were determined from displacement drug equilibrium binding assays. Vinblastine and paclitaxel could only fractionally displace [3H]-XR9576 binding, displaying Ki values significantly different from their measured Kd values. This suggests a non-competitive interaction between XR9576 and the P-gp substrates vinblastine and paclitaxel. 3 XR9576 was shown to be a potent modulator of P-gp mediated [3H]-vinblastine and [3H]-paclitaxel transport as it increased the steady-state accumulation of these cytotoxics in CHrB30 cells to levels observed in non-P-gp-expressing AuxB1 cells (EC50 = 487+/-50 nM). This inhibition of drug transport is not mediated through competition for transport since [3H]-XR9576 accumulation was not influenced by P-gp expression or function. 4 These results demonstrate that the P-gp modulator XR9576 exhibits greater selectivity, duration of inhibition and potency of interaction with this transporter than any other reported modulators. Several lines of evidence suggest that XR9576 inhibits P-gp function by binding at a site which is distinct from the site of interaction of transport substrates. The two sites may be classified as serving modulatory or transport functions.[1]
The overexpression of P-glycoprotein (P-gp) on the surface of tumor cells causes multidrug resistance (MDR). This protein acts as an energy-dependent drug efflux pump reducing the intracellular concentration of structurally unrelated drugs. Modulators of P-gp function can restore the sensitivity of MDR cells to such drugs. XR9576 is a novel anthranilic acid derivative developed as a potent and specific inhibitor of P-gp, and in this study we evaluate the in vitro and in vivo modulatory activity of this compound. The in vitro activity of XR9576 was evaluated using a panel of human (H69/LX4, 2780AD) and murine (EMT6 AR1.0, MC26) MDR cell lines. XR9576 potentiated the cytotoxicity of several drugs including doxorubicin, paclitaxel, etoposide, and vincristine; complete reversal of resistance was achieved in the presence of 25-80 nM XR9576. Direct comparative studies with other modulators indicated that XR9576 was one of the most potent modulators described to date. Accumulation and efflux studies with the P-gp substrates, [3H]daunorubicin and rhodamine 123, demonstrated that XR9576 inhibited P-gp-mediated drug efflux. The inhibition of P-gp function was reversible, but the effects persisted for >22 h after removal of the modulator from the incubation medium. This is in contrast to P-gp substrates such as cyclosporin A and verapamil, which lose their activity within 60 min, suggesting that XR9576 is not transported by P-gp. Also, XR9576 was a potent inhibitor of photoaffinity labeling of P-gp by [3H]azidopine implying a direct interaction with the protein. In mice bearing the intrinsically resistant MC26 colon tumors, coadministration of XR9576 potentiated the antitumor activity of doxorubicin without a significant increase in toxicity; maximum potentiation was observed at 2.5-4.0 mg/kg dosed either i.v. or p.o. In addition, coadministration of XR9576 (6-12 mg/kg p.o.) fully restored the antitumor activity of paclitaxel, etoposide, and vincristine against two highly resistant MDR human tumor xenografts (2780AD, H69/LX4) in nude mice. Importantly all of the efficacious combination schedules appeared to be well tolerated. Furthermore, i.v. coadministration of XR9576 did not alter the plasma pharmacokinetics of paclitaxel. These results demonstrate that XR9576 is an extremely potent, selective, and effective modulator with a long duration of action. It exhibits potent i.v. and p.o. activity without apparently enhancing the plasma pharmacokinetics of paclitaxel or the toxicity of coadministered drugs. Hence, XR9576 holds great promise for the treatment of P-gp-mediated MDR cancers.[2] In drug development, biomarkers for cerebral applications have a lower success rate compared to cardiovascular drugs or tumor therapeutics. One reason is the missing blood brain barrier penetration, caused by the tracer's interaction with efflux transporters such as the P-gp (MDR1 or ABCB1). Aim of this study was the development of a reliable model to measure the interaction of radiotracers with the human efflux transporter P-gp in parallel to the radiolabeling process. LigandTracer® Technology was used with the wildtype cell line MDCKII and the equivalent cell line overexpressing human P-gp (MDCKII-hMDR1). The method was evaluated based on established PET tracers with known interaction with the human P-gp transporter and in nanomolar concentration (15 nM). [11C]SNAP-7941 and [18F]FE@SNAP were used as P-gp substrates by comparing the real-time model with an uptake assay and μPET images. [11C]DASB [11C]Harmine, [18F]FMeNER,[18F]FE@SUPPY and [11C]Me@HAPTHI were used as tracers without interactions with P-gp in vitro. However, [11C]Me@HAPTHI shows a significant increase in SUV levels after blocking with Tariquidar. The developed real-time kinetic model uses directly PET tracers in a compound concentration, which is reflecting the in vivo situation. This method may be used at an early stage of radiopharmaceutical development to measure interactions to P-gp before conducting animal experiments.[3] |
| Molecular Formula |
C40H52N4O12S2
|
|
|---|---|---|
| Molecular Weight |
892.99
|
|
| Exact Mass |
892.287
|
|
| CAS # |
625375-83-9
|
|
| Related CAS # |
Tariquidar;206873-63-4; 625375-84-0 (mesylate);1992047-62-7 (2HCl); 625375-83-9 (methanesulfonate hydrate)
|
|
| PubChem CID |
71576652
|
|
| Appearance |
Light yellow to yellow solid
|
|
| LogP |
7.746
|
|
| Hydrogen Bond Donor Count |
7
|
|
| Hydrogen Bond Acceptor Count |
17
|
|
| Rotatable Bond Count |
11
|
|
| Heavy Atom Count |
61
|
|
| Complexity |
1130
|
|
| Defined Atom Stereocenter Count |
0
|
|
| SMILES |
S(C([H])([H])[H])(=O)(=O)O[H].S(C([H])([H])[H])(=O)(=O)O[H].O(C([H])([H])[H])C1=C(C([H])=C2C(=C1[H])C([H])([H])N(C([H])([H])C([H])([H])C1C([H])=C([H])C(=C([H])C=1[H])N([H])C(C1=C([H])C(=C(C([H])=C1N([H])C(C1C([H])=NC3=C([H])C([H])=C([H])C([H])=C3C=1[H])=O)OC([H])([H])[H])OC([H])([H])[H])=O)C([H])([H])C2([H])[H])OC([H])([H])[H].O([H])[H].O([H])[H].O([H])[H]
|
|
| InChi Key |
MNKRYFJEUQPYTI-UHFFFAOYSA-N
|
|
| InChi Code |
InChI=1S/C38H38N4O6.2CH4O3S.3H2O/c1-45-33-18-25-14-16-42(23-28(25)19-34(33)46-2)15-13-24-9-11-29(12-10-24)40-38(44)30-20-35(47-3)36(48-4)21-32(30)41-37(43)27-17-26-7-5-6-8-31(26)39-22-27;2*1-5(2,3)4;;;/h5-12,17-22H,13-16,23H2,1-4H3,(H,40,44)(H,41,43);2*1H3,(H,2,3,4);3*1H2
|
|
| Chemical Name |
N-[2-[[4-[2-(6,7-dimethoxy-3,4-dihydro-1H-isoquinolin-2-yl)ethyl]phenyl]carbamoyl]-4,5-dimethoxyphenyl]quinoline-3-carboxamide;methanesulfonic acid;trihydrate
|
|
| Synonyms |
|
|
| HS Tariff Code |
2934.99.9001
|
|
| Storage |
Powder -20°C 3 years 4°C 2 years In solvent -80°C 6 months -20°C 1 month Note: Please store this product in a sealed and protected environment, avoid exposure to moisture. |
|
| Shipping Condition |
Room temperature (This product is stable at ambient temperature for a few days during ordinary shipping and time spent in Customs)
|
| Solubility (In Vitro) |
|
|||
|---|---|---|---|---|
| Solubility (In Vivo) |
|
| Preparing Stock Solutions | 1 mg | 5 mg | 10 mg | |
| 1 mM | 1.1198 mL | 5.5992 mL | 11.1983 mL | |
| 5 mM | 0.2240 mL | 1.1198 mL | 2.2397 mL | |
| 10 mM | 0.1120 mL | 0.5599 mL | 1.1198 mL |
*Note: Please select an appropriate solvent for the preparation of stock solution based on your experiment needs. For most products, DMSO can be used for preparing stock solutions (e.g. 5 mM, 10 mM, or 20 mM concentration); some products with high aqueous solubility may be dissolved in water directly. Solubility information is available at the above Solubility Data section. Once the stock solution is prepared, aliquot it to routine usage volumes and store at -20°C or -80°C. Avoid repeated freeze and thaw cycles.
Calculation results
Working concentration: mg/mL;
Method for preparing DMSO stock solution: mg drug pre-dissolved in μL DMSO (stock solution concentration mg/mL). Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug.
Method for preparing in vivo formulation::Take μL DMSO stock solution, next add μL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O,mix and clarify.
(1) Please be sure that the solution is clear before the addition of next solvent. Dissolution methods like vortex, ultrasound or warming and heat may be used to aid dissolving.
(2) Be sure to add the solvent(s) in order.
 |
|---|
 |
 |
|
|---|
|
|